Our new paper “Translation of neutrally evolving peptides provides a basis for de novo gene evolution” has been published in Nature Ecology and Evolution on March 19 2018.
During the course of evolution, some genes are gained and others are lost. A well-established mechanism for the emergence of new genes is gene duplication. However, there is increasing evidence that some genes have not originated by gene duplication but de novo from previously non-coding regions of the genome.
The two processes can be distinguished using sequence comparisons of closely related species. In gene duplication, the new gene retains sequence similarity to the other gene copy. In contrast, genes evolved de novo show no sequence similarity to other genes. In both cases, new genes initially appear by accident. A fraction of these genes will turn out to be beneficial and be subsequently maintained by natural selection.
My interest in new genes started more than fifteen years ago. At that time, I was building a database of herpesvirus protein families at University College London. When I tried to cluster the proteins into families, some would just not cluster. These proteins had unique sequences, they did not resemble any other viral or host protein, yet they performed essential functions. Improbable as it seemed, they had to have originated from DNA sequences other than genes.
Back in Barcelona I teamed up with Jose Castresana to study gene evolution in mammals. In a paper published in 2005 we described many human and mouse proteins that lacked homologues in non-mammalian species. Following the current thinking at the time we proposed that many of them could have been generated by very rapid evolution after gene duplication. However, we also argued that it was possible that some of them had evolved de novo. The reason was that the coding sequences of the young genes were unusually small and this is something one expects for randomly occurring open reading frames but not for functional gene duplicates. Then, Macarena Toll-Riera joined the lab as a PhD student and we decided to revisit this question. With more genomes at hand, the hypothesis of de novo gene birth gained strength. The results were published in 2009 in a paper entitled Origin of primate orphan genes: a comparative genomics approach.
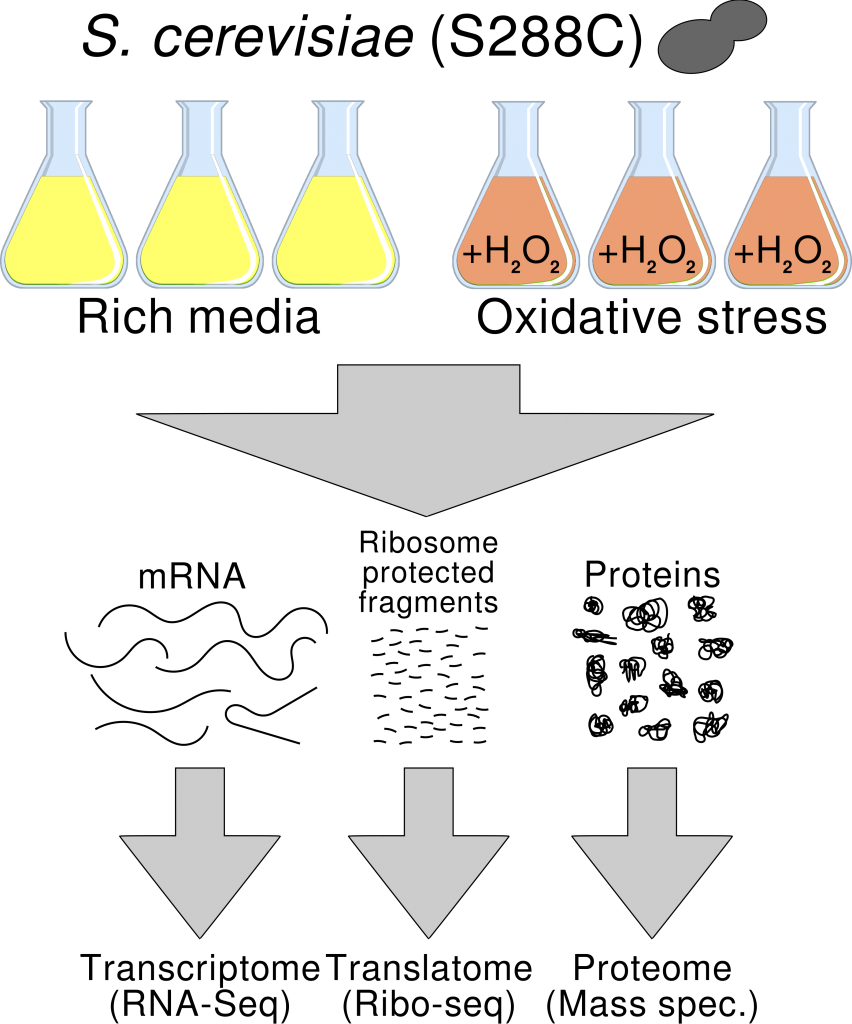
Things became exciting again when Nicholas Ingolia and co-workers reported, in 2011, widespread translation of the mouse transcriptome, including many transcripts previously believed to be non-coding. Jorge Ruiz-Orera, a new PhD in the lab, examined ribosome profiling data from different species and found clear support for the pervasive translation of the transcriptome.
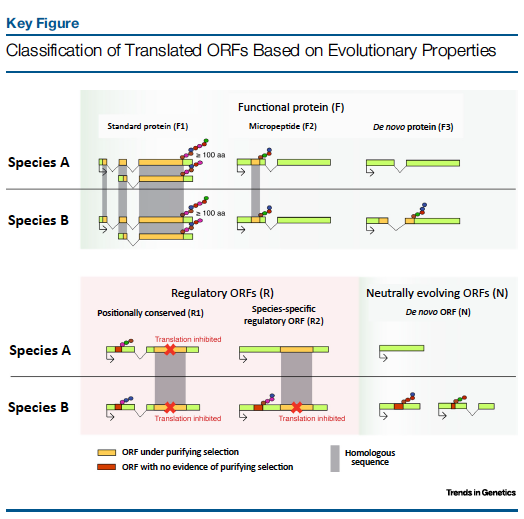
In the present study we have found that an important fraction of the translated peptides show no evolutionary conservation and evolve under no constraints. These peptides can be “tested” for new functions and eventually become new functional proteins, providing a basis for de novo gene evolution. More details of this study can be found here and in the Nature Ecology and Evolution community blog.
Mar Albà