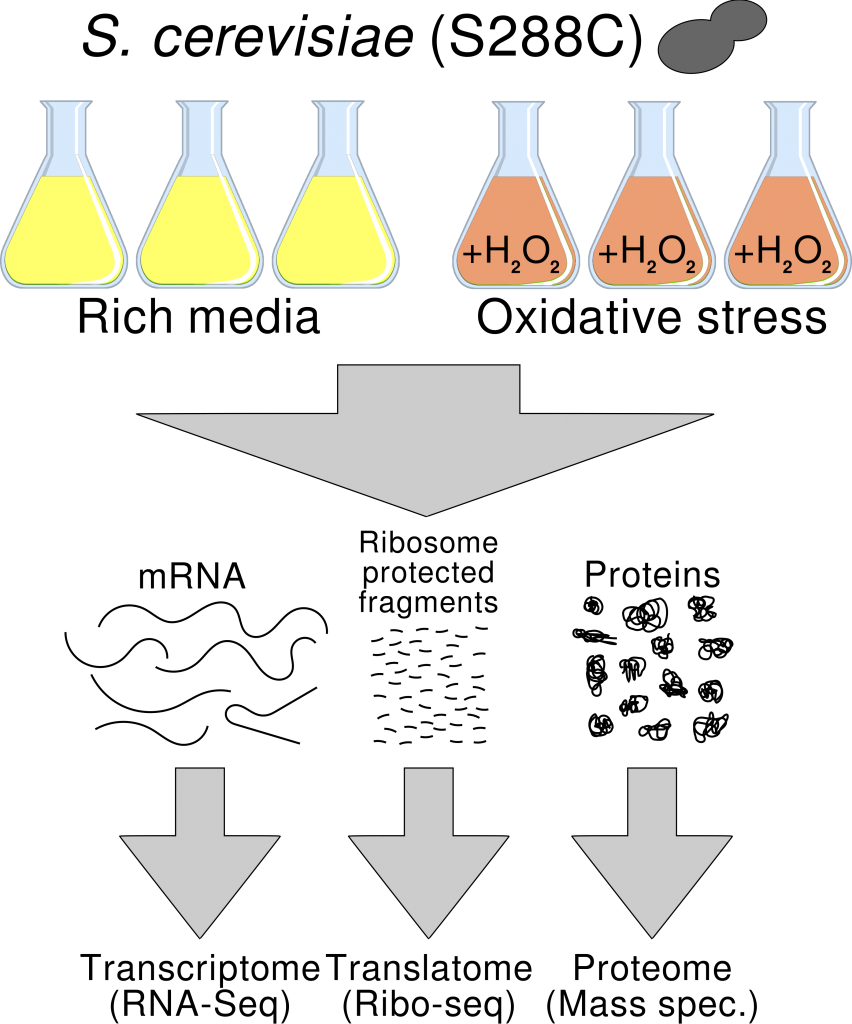
To measure changes in the expression of the genes we normally compare mRNA abundances using high throughput RNA sequencing data (RNA-Seq), as a proxy for the changes in the encoded proteins. However, it is well known that the correlation between mRNA and protein levels is far from perfect. Ideally, we would like to measure changes in the proteins themselves. The problem is that proteomics-based techniques are less sensitive, and less reproducible, than high throughput RNA sequencing. How can we get closer to the protein world while maintaining high sensitivity and specificity?
In a new study we propose to use Ribo-Seq instead RNA-Seq to perform differential gene expression analysis. Ribo-Seq is an RNA sequencing approach that specifically targets ribosome-protected RNA fragments and which shows higher correlation with proteomics data than RNA-Seq. We use this novel approach to study the response to oxidative stress in baker’s yeast. We show that the majority of genes that appear to be differentially expressed using RNA-Seq are not recovered with the Ribo-Seq-based analysis, strongly suggesting that many of these changes are not linked to changes in protein expression. This study highlights the advantages of using Ribo-Seq to understand not only translational regulation but also gene expression changes in general.
Please note that this study is now published in Scientific Reports. Follow this link to see the final version!