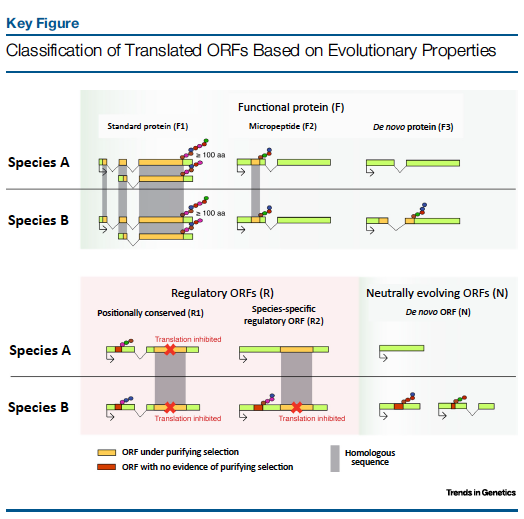
The high throughput sequencing of ribosome-protected RNA fragments, or ribosome profiling (Ribo-Seq), has uncovered the translation of thousands of novel small ORFs (< 100 amino acids) that were not annotated. These ORFs had remained hidden from annotation pipelines because of their small size, similar to that of randomly occurring ORFs in the genome. Some of these peptides show strong evolutionary conservation and have been found to play roles in development or other cellular processes. Others are located upstream of a main ORF and are translated in specific circumstances, inhibiting the translation of the main protein product.
The translation of small ORFs can also be a step towards the birth of novel protein-coding transcripts. In recent years evidence has accumulated that some protein-coding genes have originated de novo from previously non-coding genomic sequences. This requires some degree of indiscriminate transcription and translation to generate precursors. In line with this we have found that many of the mouse-specific translated small ORFs appear to evolve under no selection (Ruiz-Orera et al., 2018). This finding defies the long-held notion that any protein that is produced must be functional.
In Translation of Small Open Reading Frames: Roles in Regulation and Evolutionary Innovation (Ruiz-Orera & Albà, Trends in Genetics 2019) we review what is currently known about the small ORFome.