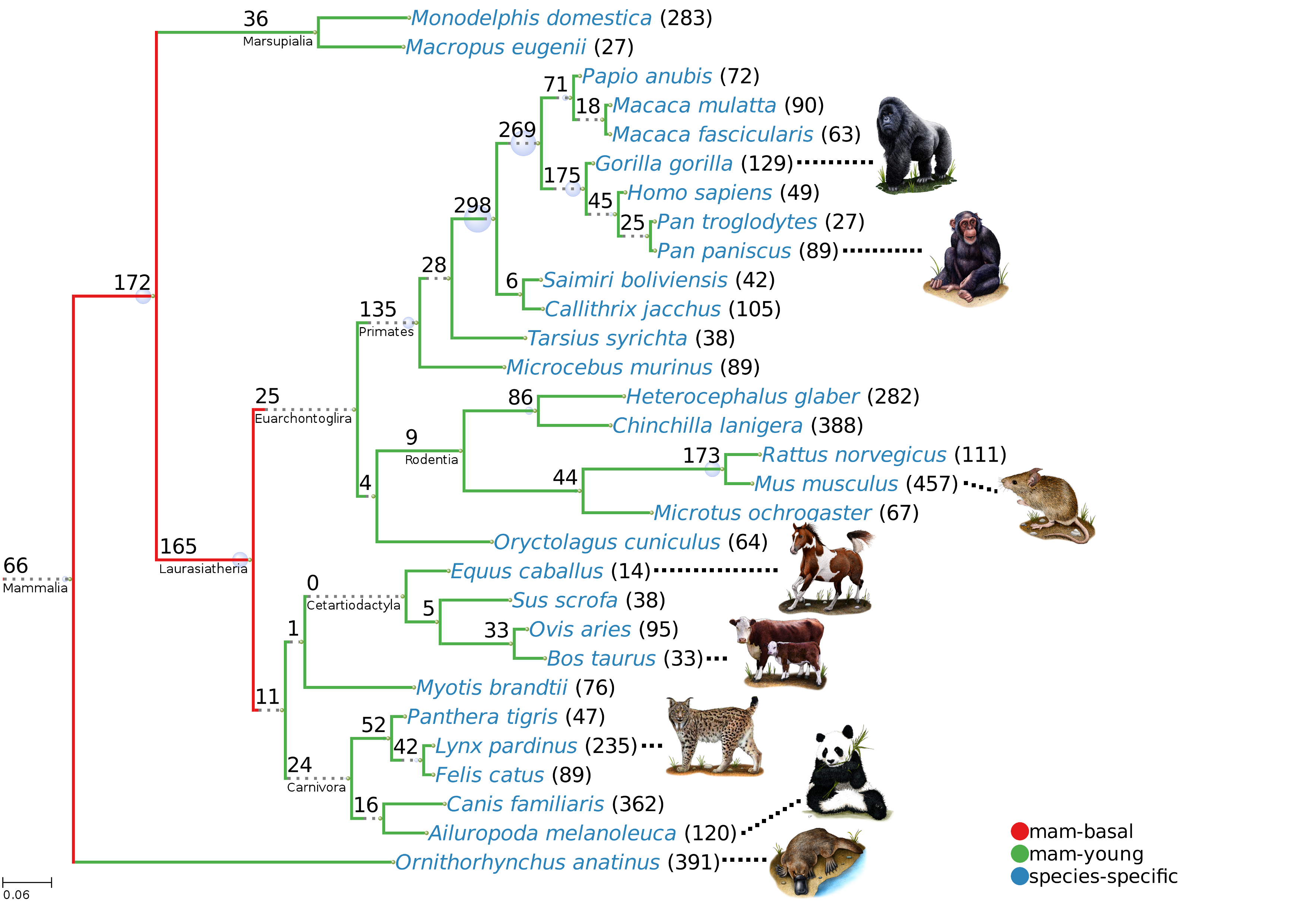
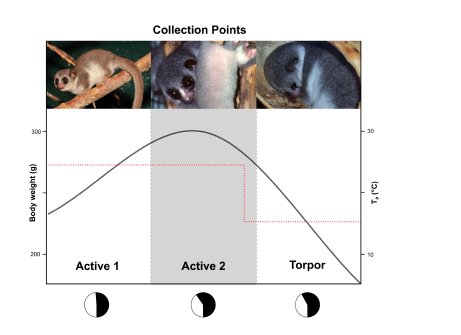
When one thinks about hibernation images of grizzly bears waking up after a long period of inaction come to mind. However, this is a more common physiological adaptation than usually thought, and many mammals hibernate, including diverse species of rats, bats, squirrels and hedgehogs. Some years ago, hibernation was also discovered in lemurs, the closest group to humans known to hibernate. The type of lemurs that hibernate are fat-tailed dwarf lemurs, which use this adaptation to endure the dry season in Madagascar.
Just before hibernation, in the so called fattening period, dwarf lemurs accumulate fat in their tails, which become very thick. This is the fuel that will allow them to survive during the several months they will spent buried in holes.
We wanted to know more about the molecular changes that took place during hibernation in lemurs. Sheena Faherty and collaborators from Duke University visited Madagascar and collected small amounts of fat tissue from the tails of the lemurs, before, during and after the hibernation period. We reconstructed the complete transcriptome from the RNA in the samples and investigated changes in gene expression during hibernation. We could detect a switch from fat storage to fat degradation, as well as inhibition of mitochondrial functions and increased protection against oxidative stress. Until recently, we knew very little about dwarf lemurs at the molecular level. Challenging as this project was, it was a joy to dive into a complete new world.
Mar Albà
The results of this study have been published in Faherty SL*, Villanueva-Cañas JL*, Blanco MB, Albà MM, Yoder AD. Transcriptomics in the wild: Hibernation physiology in free-ranging dwarf lemurs. Molecular Ecology, 29 January 2018.
link to Article