For each motif we run a sliding window along the sequences and count the number of sequences that contain at least one occurrence of the DNA motif within that window, assigning this number to the window central nucleotide. The sliding window approach allows certain degree of flexibility in the position of motifs. The size of the sliding window, which by default is 31, can be increased to detect motifs that do no have a very precise location at the cost of decreasing the significance of highly clustered motifs.
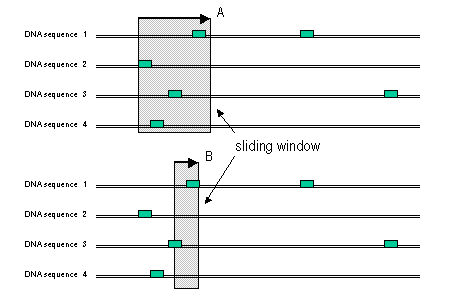 Example of motif scanning with a large (A) or small (B) window size. Small boxes represent motif matches.
Example of motif scanning with a large (A) or small (B) window size. Small boxes represent motif matches.
 Example of motif scanning with a large (A) or small (B) window size. Small boxes represent motif matches.
Example of motif scanning with a large (A) or small (B) window size. Small boxes represent motif matches.