Position-specific weight matrices (PSWM): Capture the nucleotide frecuency variation found in the columns of an aligment of binding sites.
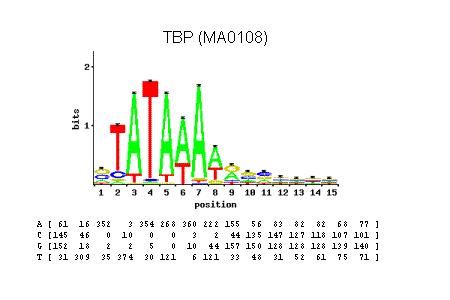 TATA-binding protein (TBP) motif represented as a logo (above) or as PSWM (below). Adapted from the JASPAR database.
TATA-binding protein (TBP) motif represented as a logo (above) or as PSWM (below). Adapted from the JASPAR database.
Available transcription factor PSWM libraries in PEAKS are from TRANSFAC (Matys et al., 2003), JASPAR (Sandelin et al., 2004) and PROMO (Messeguer et al., 2002).
A similarity cut-off is required as threshold to detect a match in the dataset. In addition a core (most conserved binding motif part) cut-off must be set to scan with TRANSFAC matrices.
Diverse matrices sub-sets are available depending on the library chosen:
| LIBRARY | SUB-SET | N PSWM |
| TRANSFAC 7 | ALL | 696 |
| vertebrate | 508 |
| plant | 78 |
| fungal | 54 |
| insect | 50 |
| nematode | 5 |
| bacteria | 1 |
| JASPAR | ALL | 123 |
| ZN-finger | 28 |
| Nuclear, NR | 12 |
| bHLH | 11 |
| bZIP | 10 |
| HOMEO | 10 |
| FORKHEAD | 8 |
| ETS | 7 |
| REL | 6 |
| TRP-cluster | 6 |
| HMG | 6 |
| MADS | 5 |
| PAIRED | 4 |
| others | 10 |
| PROMO 3 | Eukaryota | 349 |
 TATA-binding protein (TBP) motif represented as a logo (above) or as PSWM (below). Adapted from the JASPAR database.
TATA-binding protein (TBP) motif represented as a logo (above) or as PSWM (below). Adapted from the JASPAR database.